

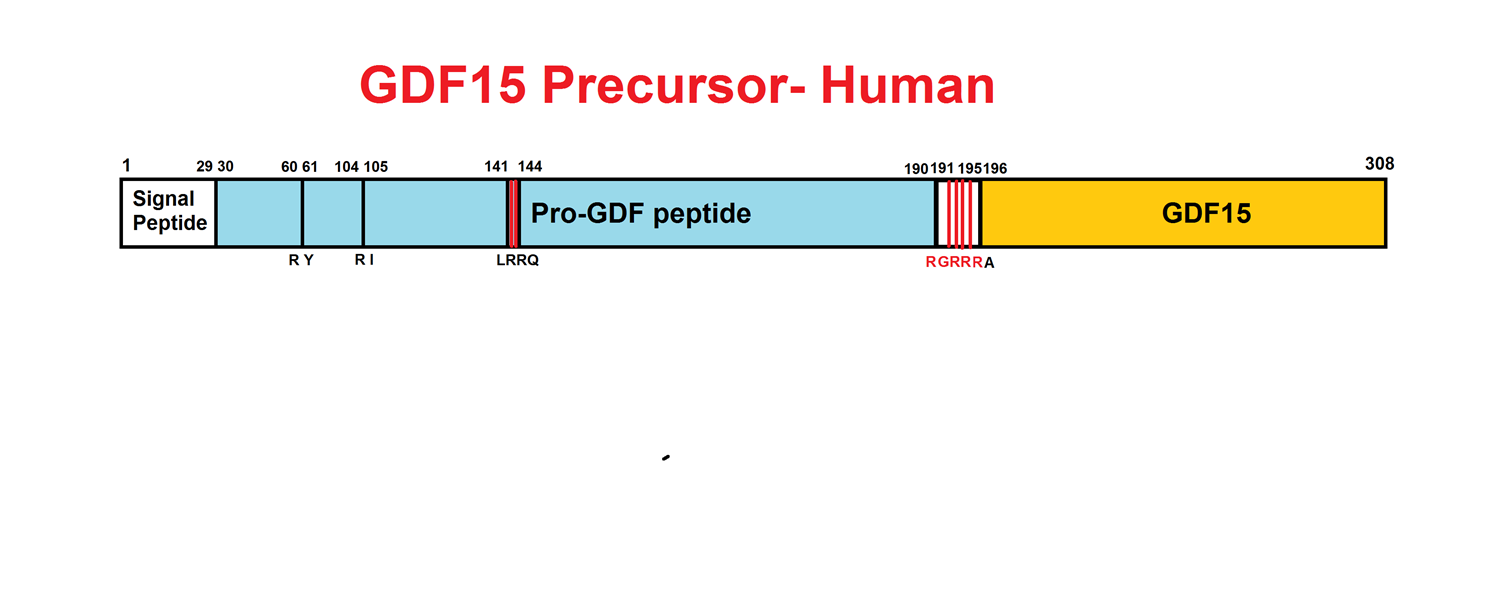
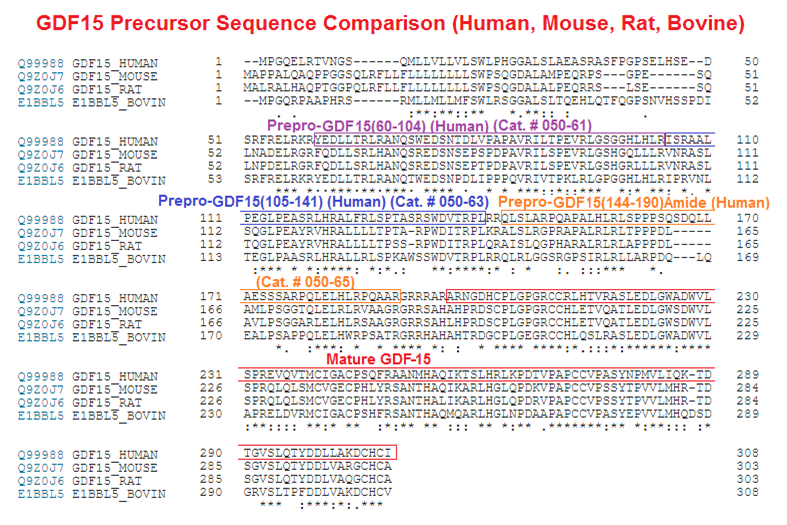
Abstract: Growth differentiation factor 15 (GDF15) is a secreted protein with pleotropic functions from the transforming growth factor β (TGF-β) family. GDF15 is synthesized as a precursor and undergoes proteolytic cleavage to generate mature GDF15. The strong appetite-suppressing effect of mature GDF15 makes it an attractive therapeutic agent/target for diseases such as obesity and cachexia. In addition, clinical studies indicate that circulating, mature GDF15 is an independent biomarker for heart failure. We recently found that GDF15 functions as a heart-derived hormone that inhibits liver growth hormone signaling and postnatal body growth in the pediatric period. However, little is known about the mechanism of GDF15 maturation, in particular the enzymes that mediate GDF15 precursor cleavage. We investigated which candidate proteases can cleave GDF15 precursor and generate mature GDF15 in cardiomyocytes in vitro and mouse hearts in vivo We discovered that three members of the proprotein convertase, subtilisin/kexin-type (PCSK) family, namely, PCSK3, PCSK5, and PCSK6, can efficiently cleave GDF15 precursor, therefore licensing its maturation both in vitro and in vivo Our studies suggest that PCSK3, -5, and -6 mediate a crucial step of GDF15 maturation through proteolytic cleavage of the precursor. These results also reveal new targets for therapeutic application of GDF15 in treating obesity and cachexia.
Li JJ, Liu J, Lupino K, Liu X, Zhang L, Pei L. Growth Differentiation Factor 15 Maturation Requires Proteolytic Cleavage by PCSK3, -5, and -6. Mol Cell Biol. 2018;38(21)
Abstract: The secretopeptidome comprises endogenous peptides derived from proteins secreted into the tumour microenvironment through classical and non-classical secretion. This study characterised the low-Mr (<3kDa) component of the human colon tumour (LIM1215, LIM1863) secretopeptidome, as a first step towards gaining insights into extracellular proteolytic cleavage events in the tumour microenvironment. Based on two biological replicates, this secretopeptidome isolation strategy utilised differential centrifugal ultrafiltration in combination with analytical RP-HPLC and nanoLC-MS/MS. Secreted peptides were identified using a combination of Mascot and post-processing analyses including MSPro re-scoring, extended feature sets and Percolator, resulting in 474 protein identifications from 1228 peptides (≤1% q-value, ≤5% PEP) - a 36% increase in peptide identifications when compared with conventional Mascot (homology ionscore thresholding). In both colon tumour models, 122 identified peptides were derived from 41 cell surface protein ectodomains, 23 peptides (12 proteins) from regulated intramembrane proteolysis (RIP), and 12 peptides (9 proteins) generated from intracellular domain proteolysis. Further analyses using the protease/substrate database MEROPS, (http://merops.sanger.ac.uk/), revealed 335 (71%) proteins classified as originating from classical/non-classical secretion, or the cell membrane. Of these, peptides were identified from 42 substrates in MEROPS with defined protease cleavage sites, while peptides generated from a further 205 substrates were fragmented by hitherto unknown proteases. A salient finding was the identification of peptides from 88 classical/non-classical secreted substrates in MEROPS, implicated in tumour progression and angiogenesis (FGFBP1, PLXDC2), cell-cell recognition and signalling (DDR1, GPA33), and tumour invasiveness and metastasis (MACC1, SMAGP); the nature of the proteases responsible for these proteolytic events is unknown. To confirm reproducibility of peptide fragment abundance in this study, we report the identification of a specific cleaved peptide fragment in the secretopeptidome from the colon-specific GPA33 antigen in 4/14 human CRC models. This improved secretopeptidome isolation and characterisation strategy has extended our understanding of endogenous peptides generated through proteolysis of classical/non-classical secreted proteins, extracellular proteolytic processing of cell surface membrane proteins, and peptides generated through RIP. The novel peptide cleavage site information in this study provides a useful first step in detailing proteolytic cleavage associated with tumourigenesis and the extracellular environment. This article is part of a Special Issue entitled: An Updated Secretome.
Greening DW, Kapp EA, Ji H, Speed TP, Simpson RJ. Colon tumour secretopeptidome: insights into endogenous proteolytic cleavage events in the colon tumour microenvironment. Biochim Biophys Acta. 2013;1834(11):2396-407.
Abstract: Macrophage inhibitory cytokine (MIC-1), a divergent member of the transforming growth factor-beta (TGF-beta)superfamily and activation associated cytokine, is secreted as a 28 kDa dimer. To understand its secretion, we examined its processing in MIC-1-transfected Chinese hamster ovary cells. Mature MIC-1 dimer arises post-endoplasmic reticulum (ER) by proteolytic cleavage of dimeric pro-MIC-1 precursor at a furin-like site. Unlike previously characterized TGF-beta superfamily members, MIC-1 dimers are also secreted in constructs lacking the propeptide. A clue to the function of the propeptide came from the observation that a range of proteasome inhibitors, including lactacystin and MG132, cause major increases in levels of undimerized pro-MIC-1 precursor. There was no effect of proteasome inhibitors on cells expressing mature MIC-1 without the propeptide, suggesting that the propeptide can signal misfolding of MIC-1, leading to proteasomal degradation. Deletion mutagenesis showed the N-terminal 28 amino acids of the propeptide are necessary for proteasomal degradation. This is the first demonstration, to our knowledge, of a quality control function in a propeptide domain of a secretory protein and represents an additional mechanism to ensure correct folding of proteins leaving the ER.Smith JS, Kreuder AJ, Dowling PM, et al. Re: "Evaluation of Enrofloxacin for Use in Cryopreservation of Zebu Bull Semen" by Ishaq et al. (Biopreserv Biobank 2019;[Epub ahead of print]; DOI: 10.1089/bio.2018.0133). Biopreserv Biobank. 2019;
Social Network Confirmation